hybrid-graph-benchmark
MUSAE Datasets
MUSAE-GitHub
Description
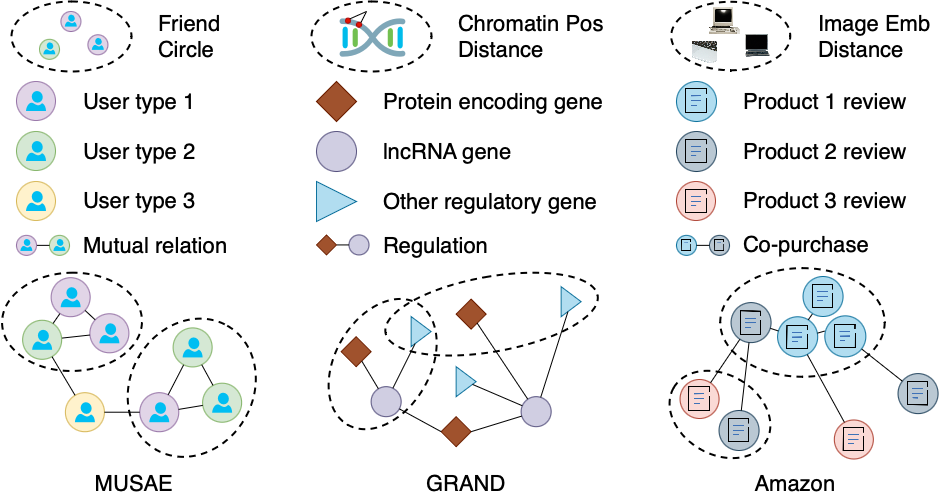
This is a large social network of GitHub developers introduced in the “Multi-scale Attributed Node Embedding” (MUSAE) paper. Nodes represent developers on GitHub, and edges are mutual follower relationships. Hyperedges are mutually following developer groups that contain at least 3 developers (i.e., maximal cliques with sizes of at least 3). The task is to predict whether a user is a web or a machine learning developer (could also be both or neither). HGB enables an option to use either the raw node features extracted based on the location, repositories starred, employer and e-mail address, or the 128-dimensional preprocessed node embeddings by MUSAE.
| #Nodes | #Edges | #Hyperedges | #Node Features | #Classes |
|---|---|---|---|---|
| 37,300 | 578,006 | 223,672 | 4,005 or 128 | 4 |
Access
Python (recommended)
from hg.datasets import GitHub
dataset = GitHub(root='/data/github')
# Data(x=[37700, 128], edge_index=[2, 578006], y=[37700], hyperedge_index=[2, 1026826], num_hyperedges=223672)
Download raw data in JSON: Zenodo
License
GNU General Public License v3.0
MUSAE-Facebook
Description
This is a large Facebook page-page network introduced in the “Multi-scale Attributed Node Embedding” (MUSAE) paper. Nodes represent verified pages on Facebook, and edges are mutual likes. Hyperedges are mutually liked page groups that contain at least 3 pages (i.e., maximal cliques with sizes of at least 3). The task is to predict which of the four categories a page belongs to: politicians, governmental organizations, television shows and companies. HGB enables an option to use either the raw node features extracted from the site descriptions, or the 128-dimensional preprocessed node embeddings by MUSAE.
| #Nodes | #Edges | #Hyperedges | #Node Features | #Classes |
|---|---|---|---|---|
| 22,470 | 342,004 | 236,663 | 4,714 or 128 | 4 |
Access
Python (recommended)
from hg.datasets import Facebook
dataset = GitHub(root='/data/facebook')
# Data(x=[22470, 128], edge_index=[2, 342004], y=[22470], hyperedge_index=[2, 2344151], num_hyperedges=236663)
Download raw data in JSON: Zenodo
License
GNU General Public License v3.0
MUSAE-Twitch
Description
These are six small Twitch user-user networks introduced in the “Multi-scale Attributed Node Embedding” (MUSAE) paper. Nodes represent gamers on Twitch, and edges are followerships between them. Hyperedges are mutually following user groups that contain at least 3 gamers (i.e., maximal cliques with sizes of at least 3). The task is to predict whether a user streams mature content. HGB enables an option to use either the raw node features extracted based on the games played and liked, location and streaming habits, or the 128-dimensional preprocessed node embeddings by MUSAE.
| Name | #Nodes | #Edges | #Hyperedges | #Node Features | #Classes |
|---|---|---|---|---|---|
| DE | 9,498 | 306,276 | 297,315 | 3,170 or 128 | 2 |
| EN | 7,126 | 70,648 | 13,248 | 3,170 or 128 | 2 |
| ES | 4,648 | 118,764 | 77,135 | 3,170 or 128 | 2 |
| FR | 6,549 | 225,332 | 172,653 | 3,170 or 128 | 2 |
| PT | 1,912 | 62,598 | 74,830 | 3,170 or 128 | 2 |
| RU | 4,385 | 74,608 | 25,673 | 3,170 or 128 | 2 |
Access
Python (recommended)
from hg.datasets import Twitch
# for other languages, use name=
dataset = Twitch('/data/twitch', name='DE') # name = ['DE', 'EN', 'ES', 'FR', 'PT', 'RU']
# Data(x=[9498, 128], edge_index=[2, 306276], y=[9498], hyperedge_index=[2, 2277625], num_hyperedges=297315)
Download raw data in JSON:
Zenodo(DE)
Zenodo(EN)
Zenodo(ES)
Zenodo(FR)
Zenodo(PT)
Zenodo(RU)
License
GNU General Public License v3.0
MUSAE-Wiki
Description
These are three Wikipedia page-page networks dataset introduced in the “Multi-scale Attributed Node Embedding” (MUSAE) paper. Nodes represent articles, and edges represent mutual hyperlinks between them. Hyperedges are mutually linked page groups that contain at least 3 pages (i.e., maximal cliques with sizes of at least 3). The task is to predict the average monthly traffic of the web page. HGB enables an option to use either the raw node features extracted based on informative nouns appeared in the text of the Wikipedia articles, or the 128-dimensional preprocessed node embeddings by MUSAE.
| Name | #Nodes | #Edges | #Hyperedges | #Node Features |
|---|---|---|---|---|
| chameleon | 2,277 | 62,742 | 14,650 | 3,132 or 128 |
| crocodile | 11,631 | 341,546 | 121,431 | 13,183 or 128 |
| squirrel | 5,201 | 396,706 | 220,678 | 3,148 or 128 |
Access
Python (recommended)
from hg.datasets import Wikipedia
# for other two graphs, use name='crocodile', 'squirrel'
dataset = Wikipedia('/data/wiki', name='chameleon') # name = ['chameleon, 'crocodile', 'squirrel']
# Data(x=[2277, 128], edge_index=[2, 62742], y=[2277], hyperedge_index=[2, 113444], num_hyperedges=14650)
Download raw data in JSON:
Zenodo(chameleon)
Zenodo(crocodile)
Zenodo(squirrel)
License
GNU General Public License v3.0
GRAND Datasets
GRAND-Tissues
Description
We select and build six gene regulatory networks in different tissues from GRAND, a public database for gene regulation. Nodes represent gene regulatory elements with three distinct types: protein-encoding gene, lncRNA gene, and other regulatory elements. Edges are regulatory effects between genes. The task is a multi-class classification of gene regulatory elements. We train a CNN and use it to take the gene sequence as input and create a 4,651-dimensional embedding for each node. The hyperedges are constructed by grouping nearby genomic elements on the chromosomes, i.e., the genomic elements within 200k base pair distance are grouped as hyperedges.
| Name | #nodes | #edges | #hyperedges | #features | #classes |
|---|---|---|---|---|---|
| Brain | 6,196 | 6,245 | 11,878 | 4608 | 3 |
| Lung | 6,119 | 6,160 | 11,760 | 4608 | 3 |
| Breast | 5,921 | 5,910 | 11,400 | 4608 | 3 |
| Artery_Coronary | 5,755 | 5,722 | 11,222 | 4608 | 3 |
| Artery_Aorta | 5,848 | 5,823 | 11,368 | 4608 | 3 |
| Stomach | 5,745 | 5,694 | 11,201 | 4608 | 3 |
Access
Python (recommended)
from hg.datasets import Grand
# name = ['Brain', 'Lung', 'Breast', 'Artery_Coronary', 'Artery_Aorta', 'Stomach']
dataset = Grand('/data/grand', name='Brain')
print(dataset[0])
# Data(x=[6196, 4608], edge_index=[2, 6245], y=[6196], hyperedge_index=[2, 15388], num_hyperedges=11878)
Download raw data in JSON:
Zenodo(Brain)
Zenodo(Lung)
Zenodo(Breast)
Zenodo(ArteryCoronary)
Zenodo(ArteryAorta)
Zenodo(Stomach)
License
GNU General Public License v3.0
GRAND-Diseases
Description
We select and build four gene regulatory networks in different genetic diseases from GRAND, a public database for gene regulation. Nodes represent gene regulatory elements with three distinct types: protein-encoding gene, lncRNA gene, and other regulatory elements. Edges are regulatory effects between genes. The task is a multi-class classification of gene regulatory elements. We train a CNN and use it to take the gene sequence as input and create a 4,651-dimensional embedding for each node. The hyperedges are constructed by grouping nearby genomic elements on the chromosomes, i.e., the genomic elements within 200k base pair distance are grouped as hyperedges.
| Name | #nodes | #edges | #hyperedges | #features | #classes |
|---|---|---|---|---|---|
| Leukemia | 4,651 | 6,362 | 7,812 | 4608 | 3 |
| Kidney_renal_papillary_cell_carcinoma | 4,319 | 5,599 | 7,369 | 4608 | 3 |
| Lung_cancer | 4,896 | 6,995 | 8,179 | 4608 | 3 |
| Stomach_cancer | 4,518 | 6,051 | 7,611 | 4608 | 3 |
Access
Python (recommended)
from hg.datasets import Grand
# name = ['Leukemia', 'Kidney_renal_papillary_cell_carcinoma', 'Lung_cancer', 'Stomach_cancer']
dataset = Grand('/data/grand', name='Leukemia')
# Data(x=[4651, 4608], edge_index=[2, 6362], y=[4651], hyperedge_index=[2, 10346], num_hyperedges=7812)
Download raw data in JSON:
Zenodo(Leukemia)
Zenodo(KidneyCancer)
Zenodo(LungCancer)
Zenodo(StomachCancer)
License
GNU General Public License v3.0
Amazon Datasets
Description
Amazon-Computers and Amazon-Photos are two e-commerce hybrid graphs based on the Amazon Product Reviews dataset. Nodes represent products, and an edge between two products is established if a user buys these two products or writes reviews for both. However, unlike other existing datasets, we introduce the image modality into the construction of hyperedge. To be specific, the raw images are fed into a CLIP classifier, and a 512-dimensional feature embedding for each image is returned to assist the clustering. The hyperedges are then constructed by grouping products whose image embeddings’ pairwise distances are within a certain threshold.
| Name | #nodes | #edges | #hyperedges | #features | #classes |
|---|---|---|---|---|---|
| Computers | 10,226 | 55,324 | 10,226 | 1000 | 10 |
| Photos | 6,777 | 45,306 | 6,777 | 1000 | 10 |
Access
Python
from hg.datasets import Amazon
# for other datasets, use name='Photos'
dataset = Amazon('/data/amazon',name='Computers') # name = ['Computers', 'Photos']
# Data(x=[10226, 1000], edge_index=[2, 55324], y=[10226], hyperedge_index=[2, 40903], num_hyperedges=10226)
License
Data Preprocessing Modules
Samplers
Following the work by GraphSAINT on simple graph sampling, we propose HypergraphSAINT, a class of hybrid graph samplers employing GraphSAINT’s graph sampling approaches. In HypergraphSAINT, we adopt the same sampling strategies in GraphSAINT for sampling the simple graph components in a hybrid graph, making three different types of samplers: HypergraphSAINTNodeSampler, HypergraphSAINTNodeSampler, and HypergraphSAINTRandomWalkSampler. As for the hyperedges, we use an intuitive procedure that any hyperedges containing at least one node in the sampled subgraph are retained, but all nodes not in the subgraph are masked out from those hyperedges. We also construct two naïve random samplers as baselines for evaluation: RandomNodeSampler and RandomHyperedgeSampler, which randomly sample a subset of nodes/hyperedges from the original hybrid graph according to a uniform sampling distribution. However, subgraphs sampled using RandomNodeSampler can be very sparse, while subgraphs sampled using RandomHyperedgeSampler can be very dense.
Example:
from hg.datasets import Facebook, HypergraphSAINTNodeSampler
# Download data to the path '/data/facebook'
data = Facebook('/data/facebook')
# Data(x=[22470, 128], edge_index=[2, 342004], y=[22470], hyperedge_index=[2, 2344151], num_hyperedges=236663)
# Create a HypergraphSAINT sampler which samples 1000 nodes from the graph for 5 times
sampler = HypergraphSAINTNodeSampler(data[0], batch_size=1000, num_steps=5)
batch = next(iter(sampler))
# Data(num_nodes=918, edge_index=[2, 7964], hyperedge_index=[2, 957528], num_hyperedges=210718, x=[918, 128], y=[918])
Data Loaders
Data Loaders can also be obtained using hg.hybrid_graph.io.get_dataset:
from hg.hybrid_graph.io import get_dataset
name = 'musae_Facebook'
train_loader, valid_loader, test_loader,data_info = get_dataset(name)
References
Ben Guebila, M., Lopes-Ramos, C. M., Weighill, D., Sonawane, A. R., Burkholz, R., Shamsaei, B., Platig, J., Glass, K., Kuijjer, M. L., and Quackenbush, J. (2022). GRAND: A Database of Gene Regulatory Network Models Across Human Conditions. Nucleic Acids Research, 50(D1):D610–D621.
Eraslan, G., Avsec, Ž., Gagneur, J., and Theis, F. J. (2019). Deep Learning: New Computational Modelling Techniques for Genomics. Nature Reviews Genetics, 20(7):389–403.
McAuley, J., Targett, C., Shi, Q., and Van Den Hengel, A. (2015). Image-Based Recommendations on Styles and Substitutes. In Proceedings of the 38th International ACM SIGIR Conference on Research and Development in Information Retrieval, pages 43–52. ACM.
Radford, A., Kim, J. W., Hallacy, C., Ramesh, A., Goh, G., Agarwal, S., Sastry, G., Askell, A., Mishkin, P., Clark, J., et al. (2021). Learning Transferable Visual Models from Natural Language Supervision. In Proceedings of the 38th International Conference on Machine Learning (ICML 2021), volume 139, pages 8748–8763. PMLR.
Rozemberczki, B., Allen, C., and Sarkar, R. (2021). Multi-Scale Attributed Node Embedding. Journal of Complex Networks, 9(2).
Zeng, H., Zhou, H., Srivastava, A., Kannan, R., and Prasanna, V. (2020). GraphSAINT: Graph Sampling Based Inductive Learning Method. In The 8th International Conference on Learning Representations (ICLR 2020). OpenReview.net.